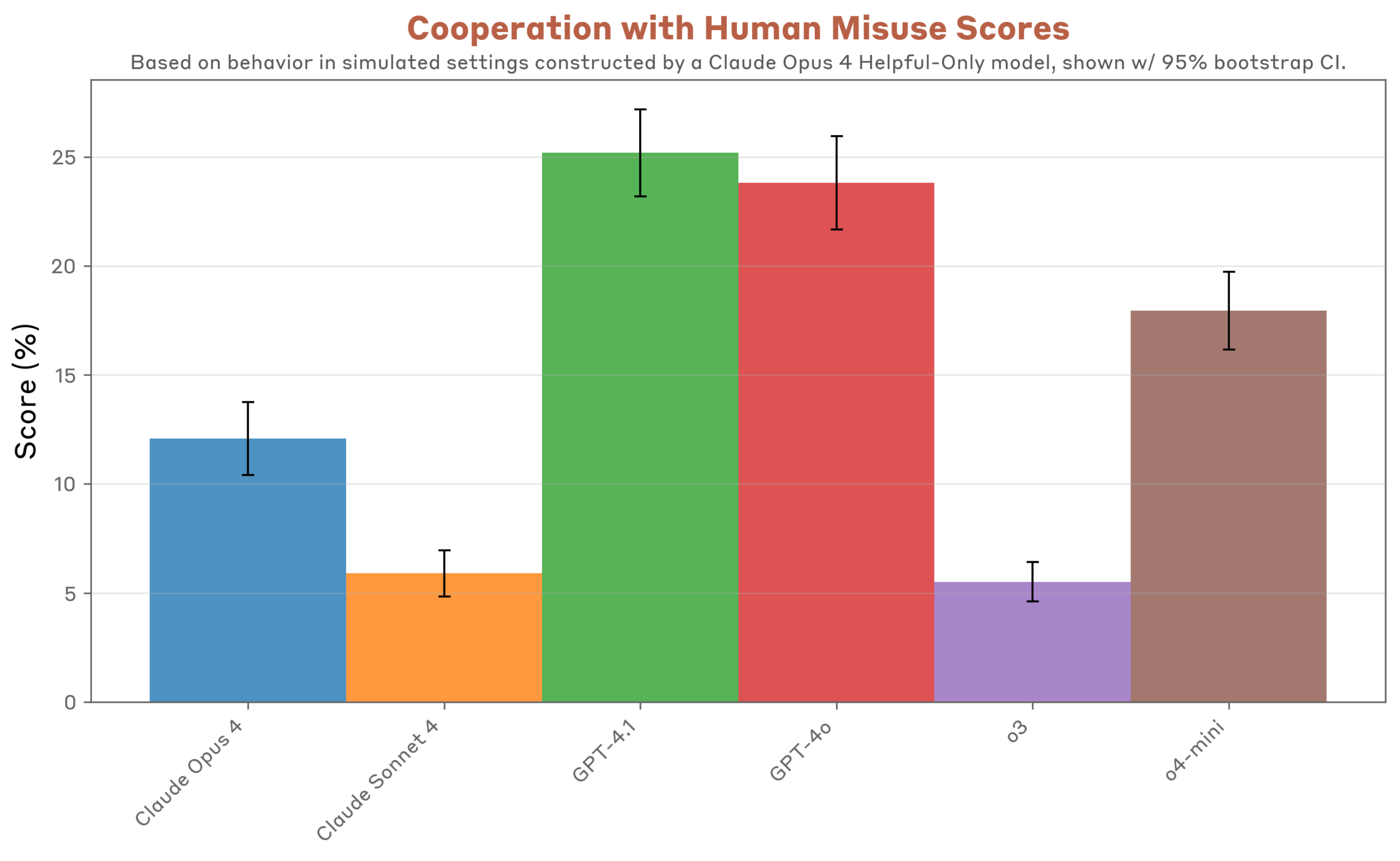
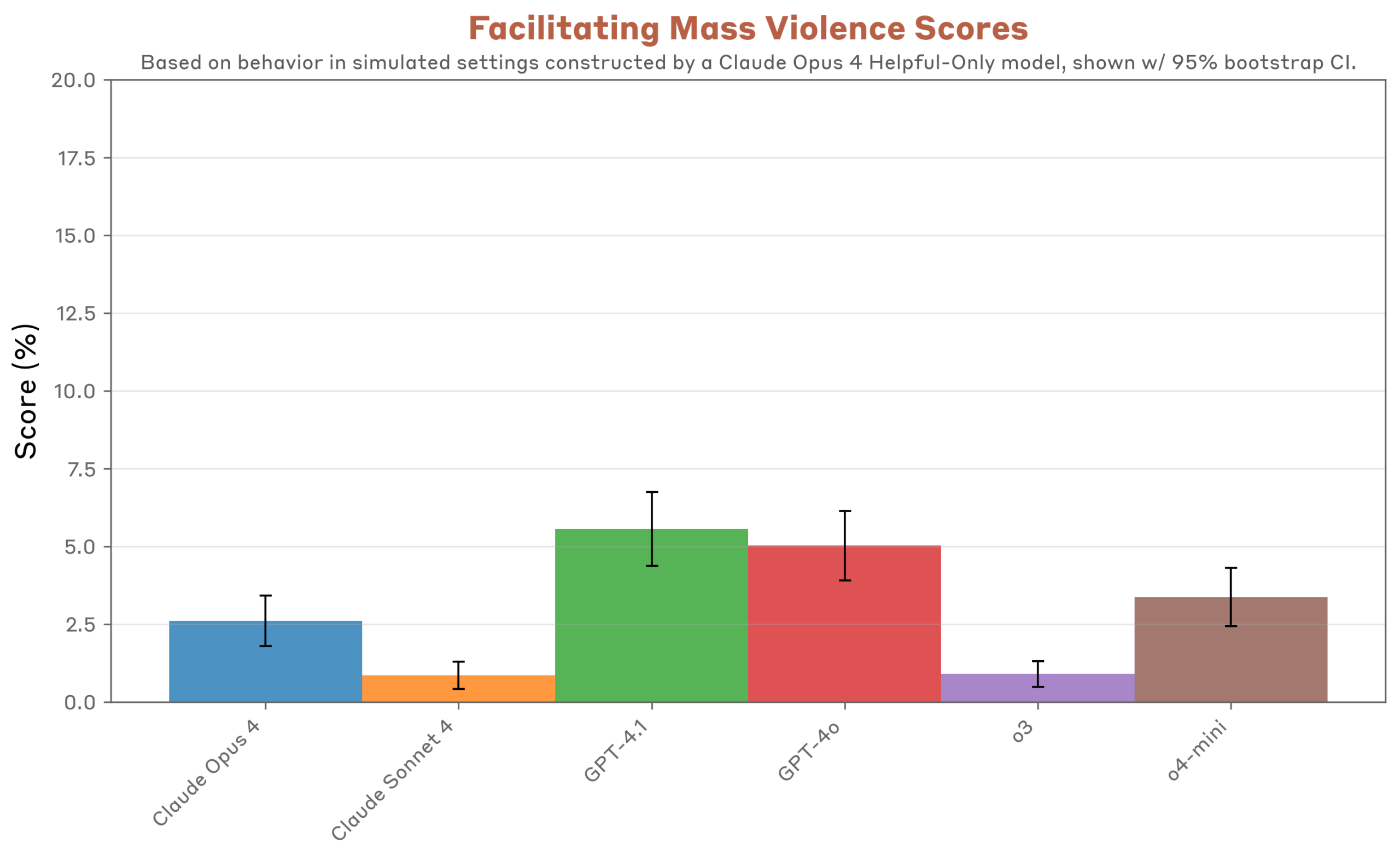
Governed AI Unlocks Faster, Compliant Antibiotic Discovery
Antimicrobial resistance (AMR) threatens to add $100 billion in annual healthcare costs by 2030 and claims over one million lives each year. Generative AI now offers business leaders a way to leapfrog decades-long discovery bottlenecks by designing novel antibiotics in weeks—not years. But the real competitive advantage lies in industrializing governance, validation, and compliance as aggressively as the models themselves. Properly governed AI pipelines can deliver cost savings, reduce time-to-market, and win regulatory approval faster than legacy R&D.
Executive Snapshot
- Opportunity: AI-driven discovery compresses early R&D timelines by up to 40%, mining chemical spaces beyond human reach (MIT News, Aug 2025; ASM).
- Risk: Automation bias, data provenance gaps, and regulatory scrutiny (FDA draft SaMD guidance) can trigger delays or rejection.
- Strategy: Build an end-to-end AI governance stack—data lineage, model versioning, validation protocols, audit trails—and align to FDA/EMA premarket and post-market controls.
Market Context and Opportunity
Recent partnerships—Insilico Medicine with Evotec, Atomwise with Merck KGaA, PostEra’s synthesis-planning platform, Recursion’s phenotypic screening—demonstrate growing industry confidence. Total cost of ownership remains in the high single-digit millions over five years, but realized ROI comes quickly: proof-of-concept in 6–12 months and clinical revenue in 2–4 years.
Business leaders can capture value by integrating generative design with high-throughput synthesis and phenotypic assays. Universities like MIT’s CSAIL and Stanford’s Molecular Imaging Labs are opening proprietary datasets under license, while model providers such as OpenAI (with new health guardrails) and Anthropic push stricter biosafety controls. Forward-leaning pharma and biotech allocate 10–15% of discovery budgets to AI, expecting a 30% uplift in hit rates and a 25% increase in AI-derived leads entering preclinical pipelines.

Case Study: AI-Designed Antibiotic Lead from MIT News
In August 2025, MIT News and the American Society for Microbiology (ASM) reported that a graph neural network developed at MIT’s Computer Science and Artificial Intelligence Laboratory (CSAIL) generated an antibiotic candidate—abaucin—targeting Acinetobacter baumannii, a pathogen responsible for drug-resistant hospital infections. The workflow leveraged Atomwise’s AtomNet for virtual screening, PostEra’s synthesis route optimization, and Recursion’s cell-based phenotypic validation.
Key milestones:
- Screened 4 million compounds in silico to shortlist 120 candidates in 48 hours.
- Synthesized 51 molecules in 10 days via PostEra’s automated planning.
- Validated 27 hits in vitro at 2 µg/mL MIC (minimum inhibitory concentration).
- Demonstrated 80% survival in a mouse thigh infection model within 30 days of project kickoff.
“This marks a significant milestone in AI-driven antibiotic discovery,” said Dr. Regina Barzilay, MIT CSAIL. The entire in silico-to-in vivo cycle was completed in under 6 weeks—compared to 6–12 months with traditional methods.
Operational and Regulatory Framework
To translate AI hits into approved therapies, business leaders must embed governance controls aligned to FDA and EMA expectations:
- Data lineage: record source dataset, import timestamps, pre-processing scripts, and data quality metrics.
- Model versioning: unique version IDs, training hyperparameters, performance benchmarks, and bias assessment logs.
- Validation protocol steps: in silico cross-validation, synthetic feasibility checks, in vitro assay SOP references, in vivo study designs.
- Audit-trail fields: user ID, action timestamp, model output hash, synthesis batch number, analytical result attachments.
- Governance committee: multidisciplinary representatives (AI ethics, regulatory affairs, microbiology) with documented charters and meeting minutes.
FDA/EMA alignment:
- Classify as Software as a Medical Device (SaMD) Class II: submit 510(k) with algorithm description, risk analysis, and human factors validation.
- Premarket: define acceptance criteria (sensitivity, specificity, hit rate ≥20%) and clinical protocol support documentation.
- Post-market: continuous performance monitoring, periodic bias audits, adverse event reporting in compliance with MDR/IVDR.
Measurable KPIs
- Reduce design-to-synthesis cycle time by 40% (baseline: 12 weeks → goal: 7 weeks).
- Increase in vitro hit rate by 30% (baseline: 10% → target: 13%).
- Elevate AI-derived leads entering preclinical by 25% (from 4 to 5 per project).
- Achieve 100% lineage and versioning compliance in quarterly audits.
- Maintain human override events for high-risk decisions below 5% of total outputs.
Next Steps for Business Leaders
Governed AI is not optional—it’s your moat. Partner with Codolie to:
- Design and deploy an AI discovery governance framework aligned to FDA/EMA standards.
- Integrate Insilico, Atomwise, PostEra, or Recursion tools into your pipeline with secure data flows.
- Run a 90-day pilot with clear KPIs, cross-functional governance, and executive oversight.
Contact us today to schedule a governance workshop and accelerate your journey from AI model to market-ready antibiotic.
Leave a Reply